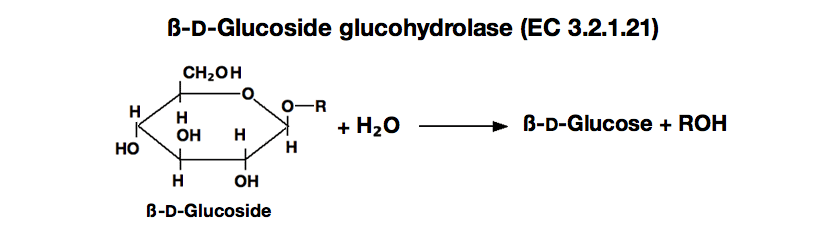ß-GLUCOSIDASE from Sweet almond
BGH-101
| Appearance: | Light yellow amorphous powder, lyophilized | ||
|---|---|---|---|
| Activity: | GradeⅠ 15U/mg-solid or more (containing approx. 30% of BSA) |
||
| Contaminant: | α-Amylase ≤5.0×10⁻⁴% |
||
| Stabilizers: | BSA, glutathione (reduced) | ||
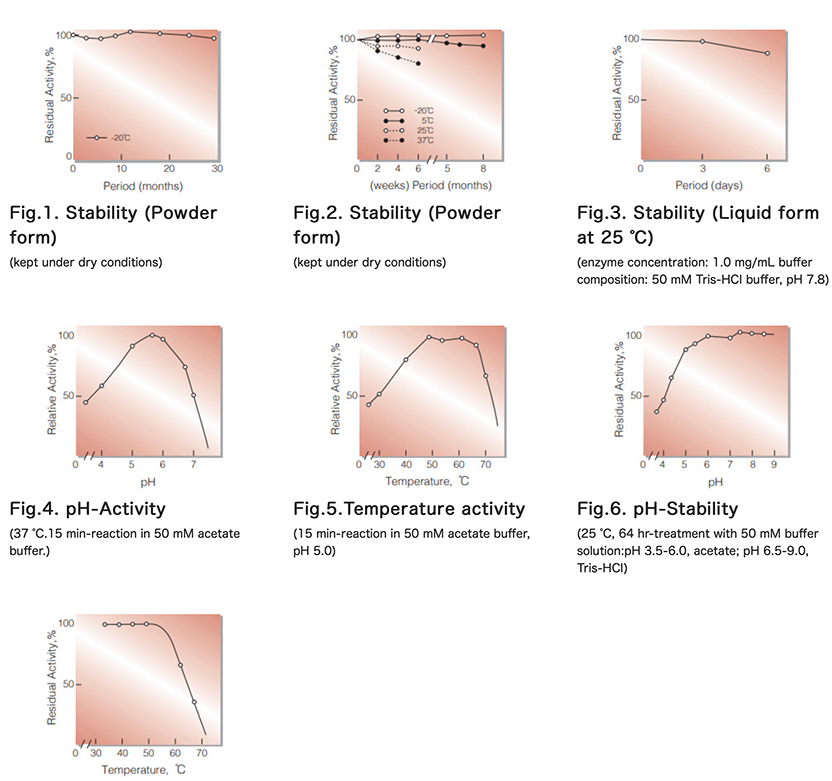
| Stability: | Stable at -20°C for at least One year(Fig.1) |
|---|---|
| Molecular weight: | approx. 110,000 |
| Isoelectric point : | 7.3 ¹ ⁾ |
| Michaelis constants : | 2.8×10⁻³M (p-Nitrophenyl-ß-D-glucopyranoside), 3.3×10⁻³M (2,4-Dichlorophenyl-ß-D-glucopyranoside) |
| Structure : | 2 subunits per enzyme molecule |
| Optimum pH : | 5.5(Fig.4) |
| Optimum temperature: | 50-55°C(Fig.5) |
| pH Stability: | pH 6.0-9.0 (25°C, 64hr)(Fig.6) |
| Thermal stability: | below 50°C (pH 7.3, 1hr)(Fig.7) |
| Effect of various chemicals: | (Table 1) |
APPLICATIONS
This enzyme is useful for structural investigations of carbohydrates and for the enzymatic determination of α-amylase in combination with α-glucosidase (AGH-211) in clinical analysis.
ASSAY
Principle:
ß-glucosidase
p-Nitrophenyl-ß-D-glucopyranoside (PNPG) ► p-Nitrophenol (PNP)+D-Glucose
The appearance of p-nitrophenol is measured at 400nm by spectrophotometry.
Unit definition:
One unit causes the formation of one micromole of PNP per minute under the conditions described below.
Method:
| A. Acetate buffer, pH 5.0 (at 25°C): | 0.1M |
|---|---|
| B. PNPG solution: | 20mM (603mg p-nitrophenyl-ß-D-glucopyranoside/100ml of H₂O)(Stable for two weeks if stored at 0-5°C) |
| C. Na₂CO₃ solution: | 0.2M (21.2g Na₂CO₃ /1,000ml of H₂O) |
| D. Enzyme diluent: | 10mM phosphate buffer, pH 7.0 containing 0.2% of BSA. |
Procedure
| Concentration in assay mixture | |
|---|---|
| Acetate buffer | 50 mM |
| PNPG | 5.0 mM |
| BSA | 0.05mg/ml |
1. Prepare the following reaction mixture in a test tube and equilibrate at 37°C for approximately 5 minutes.
1.0 ml 0.1M Acetate buffer, pH 5.0 (A)
0.5 ml Substrate solution (B)
2. Add 0.5ml of the enzyme solution* and mix.
3. After exactly 15 minutes at 37°C, add 2.0ml of Na₂CO₃ solution (C) to stop the reaction and measure the
optical density at 400nm against water (OD test).
At the same time, prepare the blank by mixing the reaction mixture with 2.0ml of Na₂CO₃ solution (C)
after incubation for 15 minutes at 37°C, followed by addition of the enzyme solution (OD blank).
* Dissolve the enzyme preparation in ice-cold 50mM Tris-HCl buffer pH 7.8 (ca. 1mg/ml) and dilute to
0.006-0.022U/ml with the enzyme diluent (D), immediately before assay.
Calculation
Activity can be calculated by using the following formula :

ΔOD (OD test−OD blank ) ×Vt × df
Volume activity (U/ml) = =ΔOD×0.0295×df
18.1×1.0 × t ×Vs
Weight activity (U/mg)=(U/ml)×1/C
- Vt
- : Total volume (4.0ml)
- Vs
- : Sample volume (0.5ml)
- 18.1
- : Millimolar extinction coefficient of p-nitrophenol under the assay condition (㎠/micromole)
- 1.0
- : Light path length (cm)
- t
- : Reaction time (15 minutes)
- df
- : Dilution factor
- C
- : Enzyme concentration in dissolution (c mg/ml)
REFERENCES
- A.K.Grover, D.D.Macmurchie and R.J.Cushley; Biochim.Biophys.Acta, 482, 98 (1977).
(Characteristics of ß-Glucosidase from almond) - R.Heyworth and P.G.Walker; Biochem.J., 83, 331 (1962).
- J.H.Hash and K.W.King; J.Biol.Chem., 232, 395 (1958).
| Chemical | Concn.(mM) | Residual activity(%) |
Chemical | Concn.(mM) | Residual activity(%) |
|---|---|---|---|---|---|
| None | − | 100 | MnCl₂ | 94.3 | |
| Metal salt | 0.5 | BaCl₂ | 93.9 | ||
| CaCl₂ |
92.7 |
FeCl₃ | 99.8 |
||
| FeSO₄ | 94.1 | o-Phenanthroline | 0.5 | 94.3 | |
| CoCl₂ | 95.5 | α,α′-Dipyridy | 0.5 | 94.3 | |
| ZnCl₂ | 95.0 | Borate | 25 | 94.1 | |
| CuSO₄ | 94.5 | PCMB | 0.05 | 94.5 | |
| HgCl₂ | 99.8 | MIA | 0.5 | 89.3 | |
| CrCl₂ | 93.9 |
NaF | 0.5 | 96.6 | |
| MgSO₄ | 96.8 | NaN₃ | 10 | 98.9 | |
| SnCl₂ | 93.6 | EDTA | 5.0 | 96.1 | |
| CdCl₂ | 93.0 | Triton X-100 | 0.5% | 102.3 | |
| AgNO₃ | 92.7 |
Na-cholate | 0.5% | 99.5 | |
| NiCl₂ | 95.5 |
PCMB, p-Chloromercuribenzoate; MIA, Monoiodoacetate; EDTA, Ethylenediaminetetraacetate.
To get a quote, contact us at info@toyobousa.com, or INQUIRY.