GLUCOAMYLASE from Rhizopus sp.
GLA-111
| Appearance: | White amorphous powder (salt-free), lyophilized | |
|---|---|---|
| Activity: | GradeⅠ 30U/mg-solid or more | |
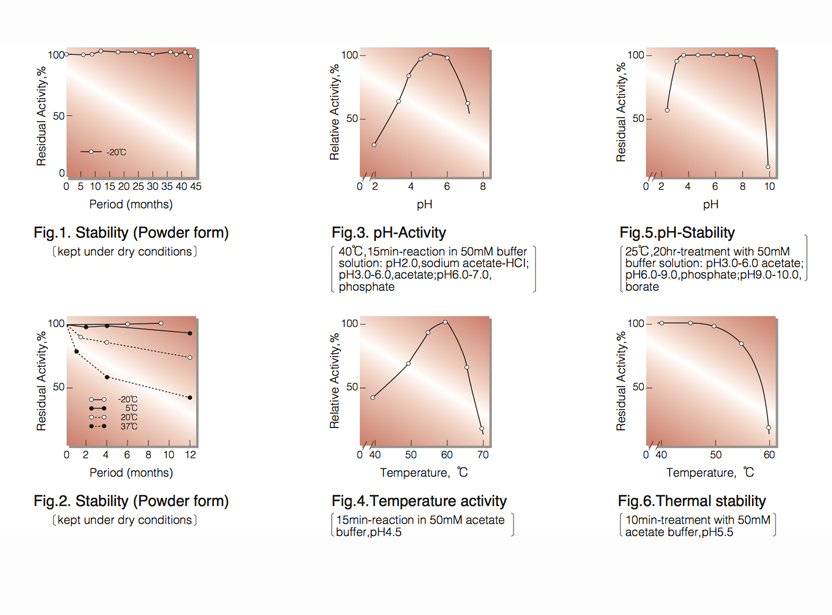
| Stability: | Stable at -20°C for at least One year (Fig.1) |
|---|---|
| Molecular weight: | approx. 70,000 ¹⁾ |
| Michaelis constants¹ ⁾: | 11±1.1×10⁻⁴M (Maltose), 3.6±0.51×10⁻⁴M (Maltotriose), 2.5±0.33×10⁻⁴M (Maltotetraose), 1.6±0.02×10⁻⁴M (Maltopentaose) |
| Structure: | |
| Optimum pH: | 4.5-5.0(Fig.3) |
| Optimum temperature: | 60℃(Fig.4) |
| pH Stability: | pH 4.0-8.5 (25℃, 20hr)(Fig.5) |
| Thermal stability: | below 45℃ (pH 5.5, 10min)(Fig.6) |
| Substrate specificty¹· ² ⁾: | This enzyme completely hydrolyzes soluble starch, amylopectin, glycogen,α-orß-limit dextrin, amylose, maltooligosaccharides and panose. |
APPLICATIONS
This enzyme is useful for structural investigation of carbohydrates and for enzymatic determination of α-amylase when coupled with the related enzymes in clinical analysis.
ASSAY
Principle:
glucoamylase
Starch+n H₂O ►n Glucose+Dextrin
The formation of glucose is measured as reducing sugar by the modified Fehling-Lehmann-Schoorl method.
Unit definition:
One unit causes the formation of ten milligrams of glucose in 30 minutes under the conditions described below.
Method:
| A. Starch solution: | 1.0%[Suspend 1.0g of soluble starch (Merck) in 90ml of H₂O, dissolve by boiling for 3min and cool down to room temperature. Add 5.0ml of 1.0M acetate buffer, pH 4.5 and fill up to 100ml with H₂O.] (Should be prepared fresh) |
|---|---|
| B. Alkaline solution: | 100g NaOH, 365g Rochelle salt・4H₂O/1,000ml of H₂O |
| C. CuSO₄ Solution: | 7.0% (70g CuSO₄・5H₂O/1,000ml of H₂O) |
| D. KI solution: | 30% (300g KI/1,000ml of H₂O)(Store in a brownish bottle) |
| E. H₂SO₄ Solution: | 25% |
| F. Na₂S₂O₃ Solution: | 50mM (49.638g Na₂S₂O₃ ・5H₂O, 4.0g Na₂CO₃ (stabilizer)/4,000ml of H₂O) (Store in a brownish bottle and keep for 3~4 days before use) |
| G. Enzyme diluent: | 10mM acetate buffer, pH 4.5 |
Procedure
| Concentration in assay mixture | |
|---|---|
| Acetate buffer | 42 mM |
| Starch | 0.8% |
1. Pipette 4.0ml of substrate solution (A) into a test tube (32ø× 200mm) and equilibrate 40℃ for about 5minutes.
2. Add 1.0ml of the enzyme solution* and mix
3. After exactly 15 minutes at 40℃, add 2.0ml of alkaline solution (B) stop the reaction.
At the same time, prepare the blank by first mixing the substrate solution with 2.0ml of alkaline solution after 15min-incubation at 40℃, followed by addition of the enzyme solution.
4. Add 2.0ml of CuSO₄ solution (C) and, after covering the test tube with a marble (40mmø) to prevent evaporation, place the test tube in a boiling water bath.
5. After 20 minutes, remove the test tube from a boiling water bath and cool down to room temperature under running water.
6. Add 2.0ml each of KI solution (D) and H₂SO₄ solution (E) in this order.
7. Shake the test tube and determine the amount of residual Cu⁺⁺ by titration with Na₂S₂O₃ solution (F).
8. Record the titers (ml) of the test (Δt) and the blank (Δb), and calculate the titration difference in ml (Δsample:Δb-Δt).
* Dissolve the enzyme preparation in ice-cold distilled water and dilute to 0.4-1.5U/ml with enzyme diluent
(G), immediately before assay.
Calculation
Activity can be calculated by using the following formula :

Weight activity (U/mg)=(U/ml)×1/C
- Δglucose
- : Titration difference (ml) for ten miligrams of glucose (Determine the titration difference by using glucose standard solution (5.0mg/ml) instead of the enzyme solution under the above assay conditions.)
- df
- : Dilution factor
- C
- : Enzyme concentration in dissolution (c mg/ml)
REFERENCES
- K.Hiromi, Y. Nitta, C.Numata and S.Ono; Biochim.Biophys.Acta, 302, 362 (1973).
- J.Fukumoto; Protein, Nucleic Acid and Enzyme, 4, 3 (1959).