PROLINE SPECIFIC ENDOPEPTIDASE from Flavobacterium sp.
PSP-101
Prolyl endopeptidase (EC 3.4.21.26)
X-Prolyl-peptide +H₂O ► X-Proline + Peptide
| Appearance: | White amorphous powder, lyophilized |
|---|---|
| Activity: | GradeⅠ 5.0U/mg-solid or more |
| Contaminants: | Leucine aminopeptidase ≤1.0×10⁻¹% Trypsin-like activity ≤1.0×10⁻¹% |
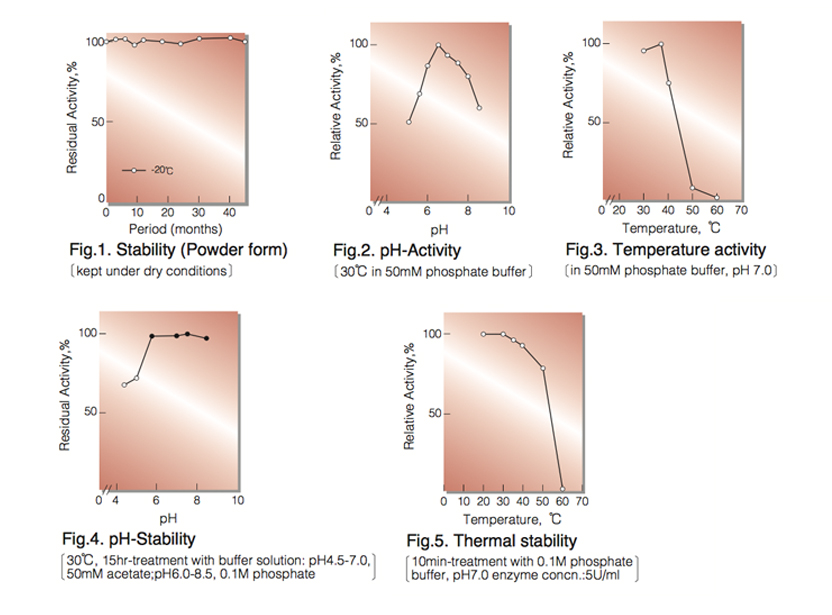
| Stability: | Stable at -20°C for at least One year (Fig.1) |
|---|---|
| Molecular weight : | approx. 78,000 |
| Isoelectric point: | 9.1 |
| Michaelis constants: | 2.5×10⁻⁵M (Z-Gly-Pro-MCA) 1.4×10⁻⁴M (Z-Gly-Pro-2NNap) |
| Structure: | Monomer |
| Inhibitors: | DFP, 3, 4-dichloroisocoumarin, Z-Gly-Pro-CH₂Cl |
| Optimum pH : | 6.5(Fig.2) |
| Optimum temperature : | 37°C (40°C) ² ⁾(Fig.3) |
| pH Stability : | 5.5-8.5 (30°C,15hr)(Fig.4) |
| Thermal stability : | below 40°C (pH7.0,10min)(Fig.5) |
| Substrate specificity: | Y-Pro(Ala)-X (Y, peptide or N-protected amino acid; X, amino acid, peptide, amide, or ester)(Table 1) |
| Effect of various chemicals : | (Table 2) |
APPLICATIONS
This enzyme is useful for the determination of amino acid sequences of peptides and proteins containing proline residues.
ASSAY
Principle:
proline specific endopeptidase
Carbobenzoxy-Gly-Pro-p-nitroanilide(Z-Gly-Pro-pNA)+H₂O ►
Z-Gly-Pro+p-Nitroaniline
The appearance of p-nitroaniline is measured at 410nm by spectrophotometry.
Unit definition:
One unit causes the formation of one micromole of p-nitroaniline per minute under the conditions described below.
Method:
| A. K-Phosphate buffer, pH 7.0: | 0.1M |
|---|---|
| B. Z-Gly-Pro-pNA solution: | 5mM[Dissolve 21.3mg of Z-Gly-Pro-pNA(MW=426.43) in ca.8ml of 40% dioxane in a hot bath at 60°C, then cool down to 25°C, fill up to 10ml with 40% dioxane](Should be prepared fresh) |
| C. Acetate buffer, pH 4.0: | 1M solution containing 10% of TritonX-100 (Store at 5°C) |
| D. Enzyme diluent: | 50mM K-phosphate buffer, pH 7.0 |
Procedure
| Concentration in assay mixture | |
|---|---|
| Phosphate buffer | 77.8 mM |
| Z-Gly-Pro-pNA | 0.926 mM |
1. Prepare the following reaction mixture in a test tube, and equilibrate at 30°C for about 5 minutes.
1.0 ml K-Phosphate buffer (A)
0.25 ml Substrate solution (B)
2. Add 0.1ml of enzyme solution* and mix by gentle inversion.
3. After exactly 5 minutes at 30°C, add 2.0ml of acetate buffer (C) to stop the reaction and measure the optical
density at 410nm against water (OD test).
At the same time, prepare the blank by first mixing the reaction mixture with 2.0ml of acetate buffer (C) after 5min-incubation at 30°C, followed by the addition of the enzyme solution (OD blank).
* Immediately before assay,dissolve the enzyme preparation in ice-cold enzyme diluent (D) and dilute to
0.05-0.2U/ml with the same buffer.
Calculation
Activity can be calculated by using the following formula :

ΔOD (OD test-OD blank)×Vt×df
Volume activity (U/ml) = = ΔOD×1.20×df
5.57×1.0× t ×Vs
Weight activity (U/mg)=(U/ml)×1/C
- Vt
- : Total volume (3.35ml)
- Vs
- : Sample volume (0.1ml)
- 5.57
- : Millimolar extinction coefficient of p-nitroaniline under the assay condition (㎠/micromole)
- 1.0
- : Light path length (cm)
- t
- : Reaction time (5 minutes)
- df
- : Dilution factor
- C
- : Enzyme concentration in dissolution (c mg/ml)
REFERENCES
- T.Yoshimoto and D.Tsuru; Agric.Biol.Chem., 42, 2417 (1978)
- T.Yoshimoto, R.Walter and D.Tsuru; J.Biol.Chem., 255, 4786(1980)
- R.Walter; Biochim.Biophys.Acta.,422, 132 (1976)
- T.Yoshimoto, R.C.Olowski and R.Walter; Biochem.J., 16, 2942 (1977)
- T.Yoshimoto, M.Fischl, R.C.Olowski and R.Walter; J.Biol.Chem., 253, 3708 (1978)
- T.Yoshimoto and D.Tsuru; Protein,Nucleic acid and Enzyme (Japanese), 29, 127 (1984)
- L.Haeffiner-Gormley, L.Parente and D.B.Wetlaufer; Int.J.Peptide Protein Res., 26, 83 (1985)
- T.Yoshimoto, K.Kawahara, F.Matsubara, K.Kado and D.Tsuru; J.Biochem., 98, 975 (1985)
| Substrate | Km (mM) |
kcat (s⁻¹) |
kcat/Km (mM⁻¹・S⁻¹) |
|---|---|---|---|
| ↓ | |||
| Pro-2NNap | not hydrolyzed | ||
| Z-Pro-2NNap | not hydrolyzed | ||
| Gly-Pro-2NNap | not hydrolyzed | ||
| Z-Gly-Pro-MCA | 0.025 | 115 | 4600 |
| Z-Gly-Pro-2NNap | 0.14 | 169 | 1212 |
| Z-Gly-Pro-pNP | 0.125 | 102 | 816 |
| Z-Ala-Pro-2NNap | 0.08 | 642 | 834 |
| Z - D -Pro-2NNap | 0.20 | 0.142 | 0.73 |
| Ala- | |||
| Z-Ala-Gly-Pro-2NNap | 0.29 | 192 | 664 |
| Z-D-Ala-Gly-Pro-2NNap | 0.14 | 38 | 271 |
| Z-Gly-Pro-Leu | 0.22 | 23 | 104 |
| Z-Gly-Pro-Phe | 0.74 | 180 | 250 |
| Z-Gly-Pro-ALa | 0.39 | 240 | 620 |
| Z-Gly-Pro-D-Ala | not hydrolyzed | ||
| Z-Gly-Pro-Leu-Gly | 0.32 | 520 | 1600 |
| Z-Gly-Pro-Leu-Ala | 0.42 | 520 | 1100 |
| Z-Gly-Pro-Leu-D-Ala | 1.5 | 1600 | 1070 |
| Z-Gly-Pro-Leu-Gly-Gly | 1.4 | 700 | 500 |
| Z-Gly-Pro-Leu-Gly-Ala | 1.82 | 1000 | 550 |
Z, Carbobenzoxy; MCA, 4-Methyl-coumaryl-7-amide; 2NNap, ß-Naphthylamide; pNP, p-Nitrophenyl ester.
| Chemical | Concn.(mM) | Residual activity(%) |
Chemical | Concn.(mM) | Residual activity(%) |
|---|---|---|---|---|---|
| None | − | 100 | NaF | 2.0 | 76.9 |
| Metal salt | 2.0 | NaN₃ | 20 | 77.3 | |
| MgCl₂ | 72.1 | DFP | 1.0 | 2.7 | |
| CaCl₂ | 74.6 | o-Phenanthroline |
2.0 | 84.9 |
|
| BaCl₂ | 74.3 |
α,α′-Dipyridyl | 1.0 | 84.9 |
|
| FeCl₃ | 52.6 | Borate | 50 | 73.4 | |
| CoCl₂ | 62.7 | IAA | 2.0 | 62.8 | |
| MnCl₂ | 68.9 |
NEM | 2.0 | 74.4 |
|
| ZnSO₄ | 56.8 | Hydroxylamine | 2.0 | 77.4 | |
| Cd(OAc)₂ | 31.9 | 3,4-Dichloroiso-coumarin | 2.0 | 9.0 | |
| NiCl₂ | 66.8 | Triton X-100 | 0.10% | 86.4 | |
| CuSO₄ | 45.2 | Brij 35 | 0.10% | 84.8 | |
| Pb(OAc)₂ | 59.8 |
Tween 20 |
0.10% | 84.1 | |
| AgNO₃ | 55.7 | Span 20 | 0.10% | 81.9 | |
| HgCl₂ | 0 | Na-cholate | 0.10% | 84.1 | |
| PCMB | 2.0 | 71.9 | SDS | 0.05% | 73.0 |
| MIA | 2.0 | 78.1 |
Ac, CH₃CO; PCMB, p-Chloromercuribenzoate; MIA, Monoiodoacetate; DFP, Diisopropylphosphorofluoridate; IAA, Iodoacetamide; NEM, N-Ethylmaleimide; SDS, Sodium dodecyl sulfate.
To get a quote, contact us at info@toyobousa.com, or INQUIRY.